DNA Content
In normal cells, errors in DNA coding trigger cell cycle arrest in the G1/S or G2/M phases and activate cell cycle-associated checkpoint pathways that mediate stage-dependent DNA repair. Once the damage has been repaired, arrested cells may either remain permanently arrested, re-enter the cell cycle, or undergo apoptosis if the damage is too severe. In cancer, the uncontrolled growth characteristic of the disease is associated with decreased time spent at DNA repair checkpoints, their complete bypass, poor or incomplete DNA repair, and accelerated proliferation in the more aggressive forms. The search for improved cancer therapies often focuses on DNA repair checkpoint restoration, which can be studied by monitoring cell cycle progression and timing.
Apart from immunophenotyping, measurement of DNA content is one of the most routinely performed flow cytometry-based assays. These experiments require the use of DNA intercalating dyes, such as 4′,6-diamidino-2-phenylindole (DAPI), propidium iodide (PI), or Vybrant™ DyeCycle™ Violet, which, when nested between DNA bases, yield signals directly proportional to the amount of DNA within the nucleus (Darzynkiewicz, Halicka, & Zhao, 2010). Since DNA content changes predictably with the stage of the cell cycle, the stoichiometric nature of the dye interactions with nucleic acids makes it possible to determine the cell cycle profile of cultured cells (Figure 1). For example, cells that are rapidly dividing have greater S-phase content in contrast to cells arrested in the G0/G1 or G2/M replication checkpoints (Darzynkiewicz et al., 2011).
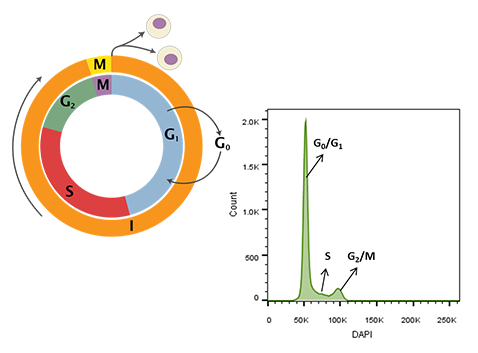
Figure 1. Cell cycle schematic (left) and DAPI-intensity histogram (right) obtained from analysis of DAPI stained single cells. DNA content doubles from G0/G1 to G2/M – the first peak in the histogram at ~50K median fluorescence intensity (MFI) is representative of G0/G1 whereas the second peak at ~100K MFI is representative of G2/M. Cells in S-phase have variable DNA content between that of cells in G0/G1 and G2/M during active DNA synthesis. Schematic representation of the cell cycle. Cytokinesis forms rapidly in the process of the cell cycle (Wheeler, 2006).
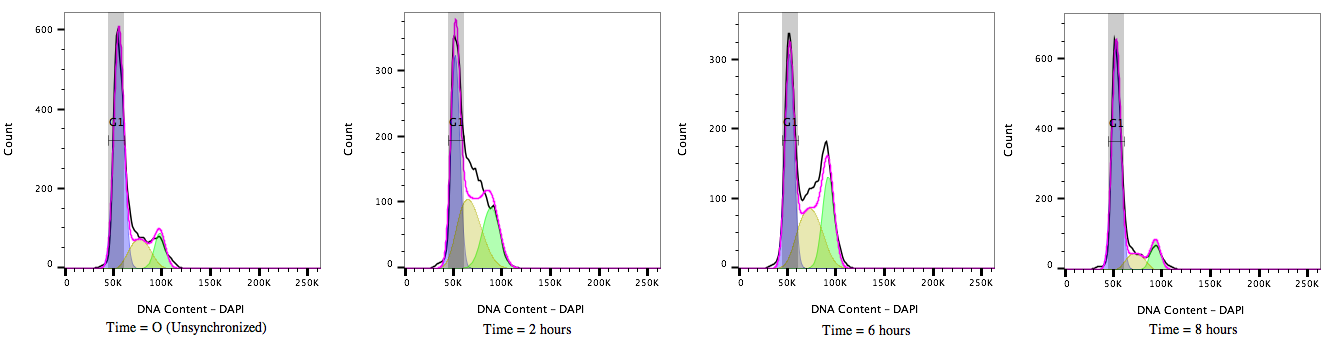
Figure 2: Cell cycle progression of HEK 293T cells treated with a cell cycle modulating agent cells and stained with DAPI (4’, 6’-Diamidino-2-Phenylindole). Over time, from T=0 to T=8 hours, the cultures move through the cell cycle transitioning out of G1/0 (A) through S-phase (B) to G2/M accumulation (C) and then cell division before resting again in the G1/0 phase (D).
Ploidy Determination
Cancer cells and cell lines often have an irregular number of chromosomes (aneuploidy), which occurs due to replication errors and uneven distribution of genetic material during mitosis. In tumors, an increased degree of aneuploidy is strongly associated with aggressive and difficult-to-treat cancers (Rancés Blanco et al., 2013; Yildirim‐Assaf et al., 2007). Ploidy determinations are crucial in characterizing tumors in the clinical setting, but they are also helpful in basic research to study cancer cell drug responses in vitro.
When measuring ploidy, DNA content is expressed as a DNA Index (DI), which is the ratio of the DNA content of the experimental sample to the DNA content of a corresponding normal control (diploid) population (Darzynkiewicz, Halicka, & Zhao, 2010). To calculate the DI correctly, an internal DNA standard must be added to each sample to ensure accurate instrument setup and DNA measurements. Chicken (CEN) and trout erythrocyte nuclei (TEN) are commonly used as internal standards since their DNA content is known to be less than that of human cells. In fact, CENs have roughly one-third the amount of genomic DNA per cell compared to humans (Iversen & Laerum, 1987), making them ideal internal standards as their DNA profiles will not overlap with those of the tested samples (see Figure 3).
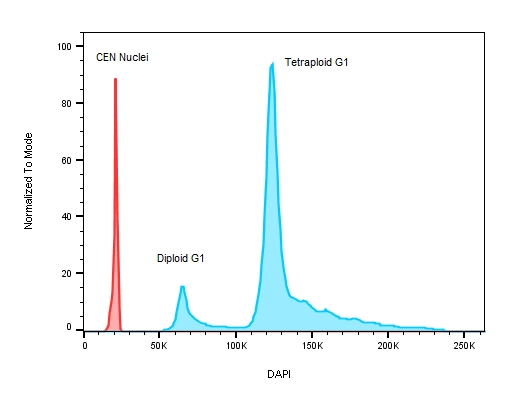
Figure 3: A plot of DNA content on a stem cell line with known replication defects causing the cells to have an abnormal DNA profile. The red peak on the histogram represents the internal control – chicken erythrocyte nuclei with lower roughly 3x less DNA when compared to human ESC test samples where the ratio between CEN to the diploid G1 peak is 3.044 and 5.767 for the tetraploid G1.
Non-proliferating human lymphocytes can serve as another useful control in the assessment of ploidy. By acquiring a patient’s own lymphocytes alongside their tumour cells, any differences between the populations can be measured (Benn & Robinson, 1996).
To obtain accurate measurements, it is recommended to collect data on isolated nuclei. Mitochondria have their own DNA and replication apparatus, and multiply in response to the increased energy demands of larger cells. As the mitochondrial burden in the cell increases, this extra-nuclear DNA can increase background DNA fluorescence, making ploidy measurements difficult (Darzynkiewicz, Halicka, & Zhao, 2010).
Benn, D. E. and Robinson, B. G. (1996). Peripheral blood mononuclear cells are a reliable internal standard for the flow cytometric DNA analysis of frozen malignant breast biopsies. Cytometry, 26, 161–165. https://doi.org/10.1002/(SICI)1097-0320(19960615)26:2<161::AID-CYTO10>3.0.CO;2-L
Darzynkiewicz, Z., Halicka, H. D., & Zhao, H. (2010). Analysis of Cellular DNA Content by Flow and Laser Scanning Cytometry. Advances in Experimental Medicine and Biology, 676, 137–147. https://doi.org/10.1007/978-1-4419-6199-0_9
Darzynkiewicz, Z., Traganos, F., Zhao, H., Halicka, H. D., & Li, J. (2011). Cytometry of DNA Replication and RNA Synthesis: Historical Perspective and Recent Advances Based on “Click Chemistry.” Cytometry. Part A : The Journal of the International Society for Analytical Cytology, 79(5), 328–337. https://doi.org/10.1002/cyto.a.21048
Iversen, O. E. and Laerum, O. D. (1987). Trout and salmon erythrocytes and human leukocytes as internal standards for ploidy control in flow cytometry. Cytometry, 8, 190–196. https://doi.org/10.1002/cyto.990080212
Rancés Blanco, Charles E. Rengifo, Mercedes Cedeño, Milagros Frómeta, and Enrique Rengifo (2013). Flow Cytometric Measurement of Aneuploid DNA Content Correlates with High S-Phase Fraction and Poor Prognosis in Patients with Non-Small-Cell Lung Cancer. ISRN Biomarkers, 2013, Article ID 354123, 8 pages. https://doi.org/10.1155/2013/354123
Wheeler, R. (2006). Schematic representation of the cell cycle. Cytokinesis forms rapidly in the process of the cell cycle illustration [Digital image]. Retrieved from https://commons.wikimedia.org/wiki/File:Cell_Cycle_2-2.svg
Yildirim‐Assaf S, Coumbos A, Hopfenmüller W, Foss H, Stein H, Kühn W. (2007). The prognostic significance of determining DNA content in breast cancer by DNA image cytometry: the role of high grade aneuploidy in node negative breast cancer. Journal of Clinical Pathology, 60(6), 649–655. https://doi.org/10.1136/jcp.2005.035550
De Novo Software. (n.d.). Chapter 6 – DNA Analysis. Retrieved from http://flowbook.denovosoftware.com/chapter-6-dna-analysis
Khan Academy. (n.d.). Cell Cycle Phases. Retrieved from https://www.khanacademy.org/science/biology/cellular-molecular-biology/mitosis/a/cell-cycle-phases
Cell Cycle Dyes
2-Dimensional cell cycle analysis combines an additional parameter, such as proliferation, with a stoichiometric DNA-binding dye to more accurately identify cells in different cell cycle stages (Darzynkiewicz and Zhao, 2014). For example, DAPI staining can be combined with fluorescently-labeled EdU or BrdU. In this assay, cells in S and G2 phases will not only have increased DAPI intensity relative to G1 cells, but also fluorescence corresponding to EdU/BrdU S-phase incorporation (see Figure 1).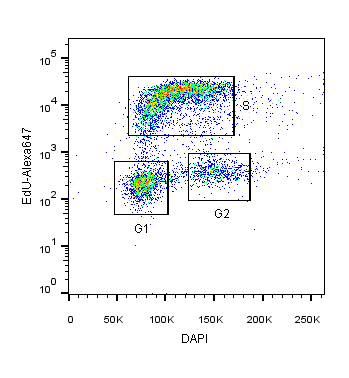
Figure 1: HEK293T cells co-stained with DAPI and EdU-Alexa 647 DNA binding dyes for the specific identification of cells in S-phase versus G1 or G2/M cell cycle phases.
Staining for markers associated with proliferation such as Ki67, phosphohistone H3, proliferating cell nuclear antigen (PCNA), topoisomerase II, and cyclin expression can be combined with a stoichiometric DNA-binding dye to increase the accuracy and resolution of cell cycle analysis.
Dyes used for DNA content analysis differ in their staining properties. Therefore, specific protocols and certain advantages/disadvantages accompany the use of each specific dye. Cell fixation and permeabilization are required for cell cycle analysis with propidium iodide (PI), 7-Aminoactinomycin D (7-AAD), 4′,6-diamidino-2-phenylindole (DAPI), and they can be achieved through various methods. Additionally, some dyes, such as PI, are not DNA-specific and will stain other double-stranded nucleic acids like RNA. When using PI, incubation of cells with RNase is necessary. In contrast, dyes like DAPI are DNA-specific and obviate this problem (Darzynkiewicz, Halicka, & Zhao, 2010).
Another important consideration when staining cells for this type of analysis is that cell concentrations must be consistent from sample to sample. This step ensures consistent saturation of DNA with the DNA-binding dye. Otherwise, issues such as peak spreading and changes in G1 peak median fluorescence intensity from sample to sample may arise, making experimental comparisons challenging. It is also important to note that the analysis of isolated nuclei may produce more accurate cell cycle characterization as opposed to the analysis of whole cells due to confounding mitochondrial DNA replication in actively proliferating cells (Darzynkiewicz, Halicka, & Zhao, 2010).
Dyes such as Hoechst33342, DRAQ5™, and Vybrant® DyeCycle enable DNA content analysis within live samples, allowing cells to be sorted by FACS for downstream assays on populations in specific cell cycle stages (Darzynkiewicz, 2011). Since these dyes are all membrane-permeant, no fixation or permeabilization step is required.
An important consideration when analyzing live stem cell populations is that drug efflux due to high expression of P-glycoprotein and other multi-drug resistance pumps can occur, preventing DNA saturation and the identification of a clear cell cycle profile. On the other hand, the dye efflux property of stem cells has been shown to be quite useful for identifying stem cells within a heterogeneous cell population through side population analysis.
The Fluorescence Ubiquitin Cell Cycle Indicator (FUCCI) system is used to track the cell cycle, and it relies on the cell-cycle dependent oscillating expression of two proteins, Geminin and Cdt1 (Sakaue-Sawano et al., 2008). The expression of these proteins is regulated via cell cycle-dependent ubiquitin-mediated proteolysis. Cdt1 expression is high during G1, whereas Geminin expression is high during the S, G2, and M phases. Thus, the expression intensity of these proteins can be detected to determine the cell cycle profile of cell populations. When non-functional variants of these proteins were linked to different fluorescent indicator proteins and expressed in HeLa cells, the detection of fluorescent signals, either from the presence of Ctd1 or Geminin, was correspondingly indicative of cell cycle stage (Sakaue-Sawano et al., 2008).
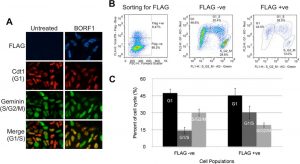 Figure 2: Screening cell cycle progression using Fucci cells. (A) Fucci cells were transfected with pMZS3F-BORF1 or left untreated and then stained for the FLAG tag on BORF1 and imaged for mKO2 (G1 cells; red), mAG (S/G2/M cells; green), and FLAG (blue). An overlay of red and green image layers revealed cells in G1/S, which display a yellow to orange color (bottom). (B) Fucci cells transfected with pMZS3F-BORF1 and stained for the FLAG tag on BORF1 were FACS sorted into FLAG-positive and FLAG-negative cells (left) and then further separated based on the expression of mKO2-Cdt1 and AG-geminin (middle and right). (C) Cell cycle stages determined from FACS analysis (as for panel B) were quantified for FLAG-positive (BORF1) and FLAG-negative cells from three independent experiments (Paladino et al., 2014).
Figure 2: Screening cell cycle progression using Fucci cells. (A) Fucci cells were transfected with pMZS3F-BORF1 or left untreated and then stained for the FLAG tag on BORF1 and imaged for mKO2 (G1 cells; red), mAG (S/G2/M cells; green), and FLAG (blue). An overlay of red and green image layers revealed cells in G1/S, which display a yellow to orange color (bottom). (B) Fucci cells transfected with pMZS3F-BORF1 and stained for the FLAG tag on BORF1 were FACS sorted into FLAG-positive and FLAG-negative cells (left) and then further separated based on the expression of mKO2-Cdt1 and AG-geminin (middle and right). (C) Cell cycle stages determined from FACS analysis (as for panel B) were quantified for FLAG-positive (BORF1) and FLAG-negative cells from three independent experiments (Paladino et al., 2014).
Darzynkiewicz, Z., Halicka, D., & Zhao, H. (2010). Analysis of cellular DNA content by flow and laser scanning cytometry. Advances in Experimental Medicine and Biology, 676, 137–147. https://doi.org/10.1007/978-1-4419-6199-0_9
Darzynkiewicz, Z. (2011). Critical aspects in analysis of cellular DNA content. Current Protocols in Cytometry, Apr, Unit 7.2. https://doi.org/10.1002/0471142956.cy0702s56
Darzynkiewicz, Z., & Zhao, H. (2014). Cell Cycle Analysis by Flow Cytometry. In eLS. John Wiley & Sons, Ltd: Chichester. https://doi.org/10.1002/9780470015902.a0002571.pub2
Darzynkiewicz, Z., Robinson, J.P. & Crissman, H.A. (Eds.). (1994). Flow Cytometry. Methods in Cell Biology, 41 & 42. Academic Press, Inc.
Hedley, D.W., Friedlander, M.I., Taylor, I.W., Rugg, C.A., & Musgrove, E.A. (1983). Method for analysis of cellular DNA content of paraffin-embedded pathological material using flow cytometry. Journal of Histochemistry & Cytochemistry, 31(11), 1333–1335. https://doi.org/10.1177/31.11.6619538
Leers, M. P., Schutte, B., Theunissen, P. H., Ramaekers, F. C., & Nap, M. (1999). Heat pretreatment increases resolution in DNA flow cytometry of paraffin-embedded tumor tissue. Cytometry, 35(3), 260–266. https://doi.org/10.1002/(SICI)1097-0320(19990301)35:3<260::AID-CYTO9>3.0.CO;2-O
Paladino, P., Marcon, E., Greenblatt, J., & Frappier, L. (2014). Identification of herpesvirus proteins that contribute to G1/S arrest. J Virol, 88(8), 4480–92. https://doi.org/10.1128/JVI.00059-14
Rabinovitch, P. S. (1994). In Darzynkiewicz, Z., Robinson, J. P., & Crissman, H. A. (Eds.), Flow Cytometry (pp. 264–296). Methods in Cell Biology, 41. Academic Press, Inc., San Diego.
Rabinovitch, P. S., Torres, R. M., & Engel, D. (1986). Simultaneous cell cycle analysis and two color surface immunofluorescence using 7-amino-actinomycin D and single laser excitation. Journal of Immunology, 136(7), 2769–2776.
Sakaue-Sawano, A., Kurokawa, H., Morimura, T., Hanyu, A., Hama, H., Osawa, H., Kashiwagi, S., Fukami, K., Miyata, T., Miyoshi, H., Imamura, T., Ogawa, M., Masai, H., Miyawaki, A. (2008). Visualizing Spatiotemporal Dynamics of Multicellular Cell-Cycle Progression. Cell, 132(3), 487–498. https://doi.org/10.1016/j.cell.2007.12.033
Vindeløv, L. L., Christensen, I. J., & Nissen, N. I. (1983). A detergent-trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry, 3(4), 323–327. https://doi.org/10.1002/cyto.990030503
Watson, J. V., Chambers, S. H., & Smith, P. J. (1987). A pragmatic approach to the analysis of DNA histograms with a definable G1 peak. Cytometry, 8(1), 1–8. https://doi.org/10.1002/cyto.990080101
Clonetech. (n.d.). Fluorescent proteins and reporters – Fucci cell cycle vectors. https://www.takarabio.com/products/cell-engineering-and-gene-regulation/cell-cycle-research/fucci-cell-cycle-vectors
DeNovo Software. (n.d.). DNA analysis. In Flowbook. http://flowbook.denovosoftware.com/chapter-6-dna-analysis
Thermo Fisher Scientific. (n.d.). Premo™ Fucci cell cycle sensor (BacMam 2.0). https://www.thermofisher.com/order/catalog/product/P36239
Learn More

Work Around Very Talented People
Meet the friendly and diverse team that keeps the University of Toronto Flow Cytometry Facility running on a day-to-day basis.

