Cellular replication is a continuous process in all biological systems where tissues undergo constant renewal. However, most cells are quiescent and terminally differentiated, existing in a permanent resting phase where they neither divide nor prepare to divide. When cells become cancerous, somatic mutations in the genome bypass the control mechanisms that normally keep quiescent cells in check, allowing them to proliferate without constraint.
The use of fluorescent probes and flow cytometry to study proliferation can provide valuable insights into the effects of drug treatments on cancer cells and other cellular models. This method can monitor various subcellular events, such as gene activation or silencing, protein expression or degradation, DNA replication, and cell cycle arrest. In addition, the number of cell divisions occurring in culture can be counted to estimate replication rates. Unlike other techniques, these assays are not limited to homogeneous samples and can be combined with several markers to study distinct populations within heterogeneous mixtures for differential analyses. Below are some details on some of the more popular flow cytometry-based proliferation experiments.
Carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) incorporation for proliferation and cell tracking
As a cell prepares to divide, its membrane enlarges and then is split evenly between its two daughter cells. Carboxyfluorescein diacetate succinimidyl ester (CFDA-SE) and other similar proliferation dyes: PKH-26 (Sigma-Aldrich: PKH26GL), CellTrace™ Violet (Thermo Fisher Scientific Inc.: C34557) take advantage of this biology allowing researchers to count the number of cell divisions that take place in culture. These dyes passively diffuse across the plasma membrane, become fluorescent upon the cleavage of their diacetyl groups by intracellular esterases and are covalently linked to intracellular proteins allowing their long term retention within the cell. As cells then divide, the amount of fluorescent label present within each division cohort is diminished by half, compared to the parent, until no/very little carboxyfluorescein succinimidyl ester (CFSE) can be detected in the cells. When plotted on a histogram (Figure 1), the number of divisions may be counted, however for more detailed analyses this data should be run through modeling software.

Figure 1: Human peripheral blood mononuclear cells were stained with BioLegend’s CFSE Cell Division Tracking Kit (Cat # 423801), and then stimulated with (filled histogram) or without (open histogram) phytohemagglutinin-A (PHA) for 5 days. On day 5, cells were harvested and the CFSE fluorescent staining was analyzed by flow cytometry. BioLegend. (2023). CFSE Cell Division Tracker Kit. Retrieved from https://www.biolegend.com/en-us/products/cfse-cell-division-tracker-kit-9396?pdf=true&displayInline=true&leftRightMargin=15&topBottomMargin=15&filename=CFSE%20Cell%20Division%20Tracker%20Kit.pdf&v=20230114013553
Roederer, M. (2011). Interpretation of cellular proliferation data: Avoid the panglossian. Cytometry, 79A, 95–101. https://doi.org/10.1002/cyto.a.21010
Two–Dimensional Cell Cycle Analysis for Proliferation:
BRDU/EDU INCORPORATION:
When cells progress through the cell cycle, accurately determining the proportion of cells in S-phase with just a single parameter can be difficult. This is especially true since DNA content analyses rely on one of two algorithms:
- Dean-Jett-Fox (DJF), accounts for abnormally distributed cell cycle phases
- Watson Pragmatic, assumes a Gaussian distribution for G0/G1 and G2/M phases
to calculate the proportion of cells in each cell cycle stage. However, these algorithms are imperfect because there is often some error in determining the true percentage of cells in each phase due to overlap in the G1/S and S/G2/M phase transitions, depending on how well the analytes fit the models.
Bromodeoxyuridine (BrdU) is a DNA base analogue that, like 3H-thymidine, can be incorporated into newly synthesized DNA when added to cell cultures. When cells are probed with a fluorescent anti-BrdU antibody, a more accurate representation of the cells in each of the cell cycle phases can be obtained and compared over the course of treatments. Changes in the number of cells in S-phase can indicate accelerated/decelerated cell division when compared to the control. In the figure below, excerpted from J. Virol. 2014;88:4480–4492, a clear difference in the percentage of cells found in S-phase is observed between the two methods, where the BrdU method yields a greater value in both the FLAG -ve and FLAG +ve sorting conditions.
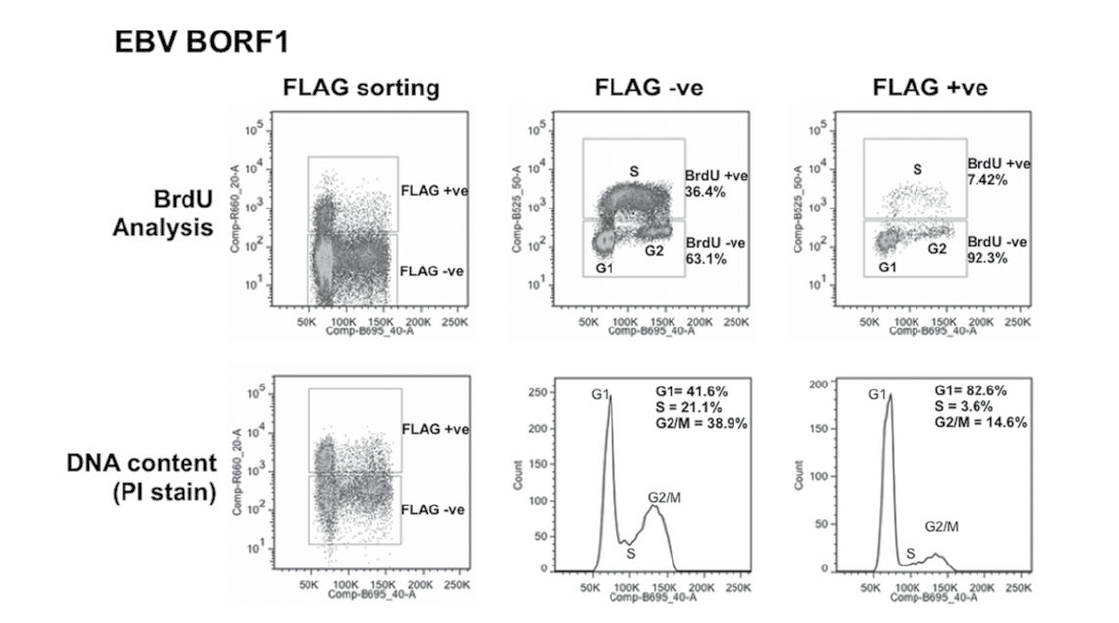
Figure 2: Cell cycle effects of EBV proteins in CNE2Z cells. CNE2Z cells were transfected with plasmids expressing FLAG-tagged BORF1 and 24 hrs later, were either pulse labelled with BrdU (top) or harvested directly and stained with propidium iodide (PI). Cells were divided into FLAG-positive and FLAG-negative cells by FACS and then analyzed for BrdU (S phase) and PI (DNA content) in the top panels or PI alone (bottom).
A newer and improved version of the BrdU assay is the EdU Click-iT protocol, which was developed by Thermo Fisher Scientific Inc. The traditional BrdU assay requires harsh denaturing conditions to expose the BrdU binding sites to antibodies, making it challenging to combine with other probes. In comparison, the EdU Click-iT protocol is less harsh, quicker, and provides a viable alternative to BrdU.
Paladino, P., Marcon, E., Greenblatt, J., & Frappier, L. (2014). Identification of herpesvirus proteins that contribute to G1/S arrest. Journal of virology, 88(8), 4480–4492. https://doi.org/10.1128/JVI.00059-14
Wallace, P. K., Tario, J. D., Fisher, J. L., Wallace, S. S., Ernstoff, M. S., & Muirhead, K. A. (2008). Tracking antigen-driven responses by flow cytometry: Monitoring proliferation by dye dilution. Cytometry, 73A, 1019–1034. https://doi.org/10.1002/cyto.a.20619
Learn More

Work Around Very Talented People
Meet the friendly and diverse team that keeps the University of Toronto Flow Cytometry Facility running on a day-to-day basis.

